

🔬 Research Summary by Rakshith Sharma Srinivasa, who obtained his PhD from Georgia Institute of Technology in 2020 and his research interests include multimodal machine learning, compressed sensing and high dimensional statistics.
[Original paper by Rakshith Sharma Srinivasa, Cheng Qian, Brandon Theodorou, Jeffrey Spaeder, Cao Xiao, Lucas Glass, and Jimeng Sun]
Overview: Effective patient enrollment for clinical trials in healthcare requires the recruitment of a cohort of patients who are both eligible for the trial and whose population characteristics reflect that of the overall population. This paper develops a machine learning algorithm to select a set of trial sites for patient recruitment that collectively meet the above criteria. The algorithm is trained to account for patient eligibility, trial site quality, and patient diversity using health insurance claims, part clinical trials performance data, and census data and identifies trial sites that maximize both patient enrollment and patient diversity.
Introduction
The development of vaccines during the COVID-19 pandemic demonstrated the importance of efficient and reliable clinical trials. While some aspects of reliability can be addressed by designing effective methods for analyzing the trial results, it is also crucial to ensure that the study population (the cohort of patients recruited for the clinical trial) matches that of the overall population. This enhances the generalizability of the results of the study. Specifically, the demographic characteristics of the study population in terms of race and ethnicity must reflect that of the overall population. This has been further emphasized by the U.S. Food and Drug Administration (FDA), which can issue multiple guidelines over the past few years to enhance racial and ethnic diversity in the study population. Addressing patient diversity in clinical trials is now a key area of research among the various stakeholders in the clinical trials industry.
In this work, we aim to develop a machine learning algorithm to identify trial sites for a given clinical trial that helps maximize patient enrollment and diversity. We utilize a rich, multimodal dataset consisting of insurance claims from the trial sites, investigator performance from trials conducted at the trial sites, and population demographics using census data to rank the trial sites under consideration regarding patient eligibility and patient diversity. We find that our algorithm consistently improves the diversity of the enrolled patient cohort while preserving the number of eligible patients enrolled.
Key Insights
Ranking sites and investigators for clinical trials
Clinical trials often span multiple years of study and involve careful study designs, patient outreach and recruitment, conducting the trial, and analyzing the resulting data. For any new drug or clinical intervention that needs to be studied, it is a common practice for drug developers to hire a contract research organization (CRO) to identify the investigators and the trial sites to conduct the study. Designated investigators at the trial sites use their medical expertise and patient access to help recruit patients and conduct studies.
Hence, choosing the right set of trial sites and investigators often becomes crucial in reaching patient enrollment goals. This process of selecting a set of sites to conduct the study is known as trial-site matching and is inherently a ranking process. It is still common that this process is conducted manually or using very basic matching between a trial’s target condition (or disease) and the investigators’ treatment history statistics. However, it is our belief that machine learning is a better tool to process large-scale patient data from the sites, the performance of the investigators from past clinical trials, and patient demographics data at the location. This paper builds upon this insight to develop an algorithm to rank sites for a given clinical trial.
Interpreting trial-site matching as a machine learning task
A clinical trial is described in terms of a set of inclusion and exclusion criteria that provide specific requirements to be met by the patients enrolled in the trial. Several examples can be found on the clinicaltrials.gov website. For a given trial and a list of candidate sites, trial-site matching entails matching the criteria to the set of patients accessible at the site and checking for the availability of experienced investigators conducting the required studies. Further, patient diversity in terms of race and ethnicity also needs to be determined. The main underlying challenges are the following:
1. Patient eligibility needs to be predicted indirectly from their medical history. While specific information related to the trial inclusion and exclusion criteria may not be explicitly available, they might need to be uncovered using information from their past hospital visits.
2. Investigators’ ability to enroll and retain eligible patients during the entire study duration needs to be predicted using information about the patients they have access to and their performance from past studies that they were part of.
3. The expected patient diversity in the set of recruited patients needs to be predicted using the local population’s demographics at the site.
The above challenges can be modeled as a prediction task using relevant data. For patients’ medical history, we use a proprietary insurance claims dataset that captures the medical history of patients who visited the trial sites. For investigators’ past performance in clinical trials, we use a dataset containing the number of patients at the beginning and the end of clinical trials that the investigators conducted in the past. For patient population demographics at the sites, we use the race and ethnicity information from the US census data available at https://statisticalatlas.com. Our paper provides more details on how these datasets were processed further and how the features to train the model were extracted. Finally, the machine learning task can be described as using the clinical trial description, patient, and investigator features from a list of candidate sites to predict and rank the sites in terms of the expected patient enrollment and diversity.
Findings
The complex interaction between a clinical trial’s criteria, patients’ medical history, investigators’ experience, and patient demographics makes machine learning a very useful tool for solving the problem of trial-site matching. In our experiments, we trained a machine learning model using data from 3480 clinical trials and tested the trained model on a test set of 480 trials that were not part of the training data. The model is fed with the trial description in text and investigator and patient data corresponding to a list of candidate sites for each trial. The model then ranks the sites to maximize patient enrollment and diversity. We then consider the top 10 sites identified by the model and measure the resulting patient diversity and the number of patients eligible at the site to enroll in the study.
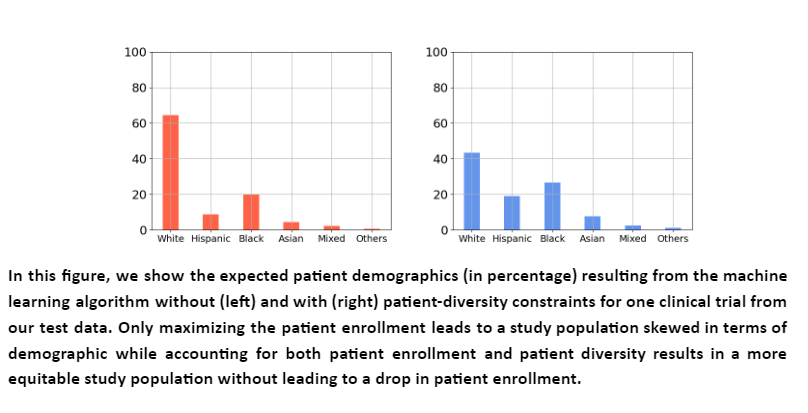
The model successfully identified sites and investigators for each trial such that the resulting patient population was diverse and equitable to racial subgroups on average. Further, the expected patient enrollment from the sites did not reduce when diversity was considered. This shows a real opportunity to improve patient diversity in clinical trials without affecting patient enrollment. We show below an illustration of one particular trial from the test set. Particularly, patient demographics obtained using algorithms that do not consider patient diversity (red) are compared to that of our algorithm (blue).
Between the lines
Clinical trial outcomes can impact different population subgroups differently if all the subgroups are not accounted for during the trial. Currently, efforts are underway in the clinical trials industry to address this concern. Our research findings indicate that machine learning can be a great tool to process the complex interactions between the various aspects affecting trial outcomes, such as patient access, investigator experience, and population demographics. We hope that this problem gets more attention from the machine learning community.
